---
title: "Exploratory Data Analysis"
---
```{r}
#| label: set-up
#| code-fold: true
#| code-summary: "Check out my code"
#| warning: false
#| message: false
# Packages ---------------------------------------------------------------------
pacman::p_load(
dplyr, # For data manipulation
tidyr, # For even more data manipulation
stringr, # String manipulation
ggplot2, # For cool plots
ggalluvial # For alluvial diagrams
)
# Functions --------------------------------------------------------------------
func_files <- c(
here::here("R/count_transitions.R"),
here::here("R/extract-years.R"),
here::here("R/plot_cognitive_scores.R"),
here::here("R/plot_transitions.R")
)
purrr::walk(func_files, source)
# Theme ------------------------------------------------------------------------
colour = "#212427"
theme_clean <- function() {
theme_minimal() +
theme(
plot.title = element_text(hjust = 0.5, size = 14, face = "bold", colour = colour ),
plot.subtitle = element_text( hjust = 0.5, size = 10, colour = colour),
axis.title = element_text(size = 10,face = "bold",colour = colour),
strip.text = element_text(size = 10,face = "bold",colour = colour),
panel.grid = element_blank(),
)
}
# Read in data -----------------------------------------------------------------
data <- read.csv(here::here("analysis/data/data.csv"))
```
Our updated dataset contains 13 columns of cognitive classifications spanning from 1996 to 2022. These columns reflect dementia status across each wave, using the same criteria and structure as the original Langa-Weir dataset, now extended to include the most recent 2022 wave.
```{r}
#| label: count-transitions
dementia_data <- data |>
count_transitions(years = seq(1996, 2022, by = 2))
glimpse(dementia_data)
```
## Missing data
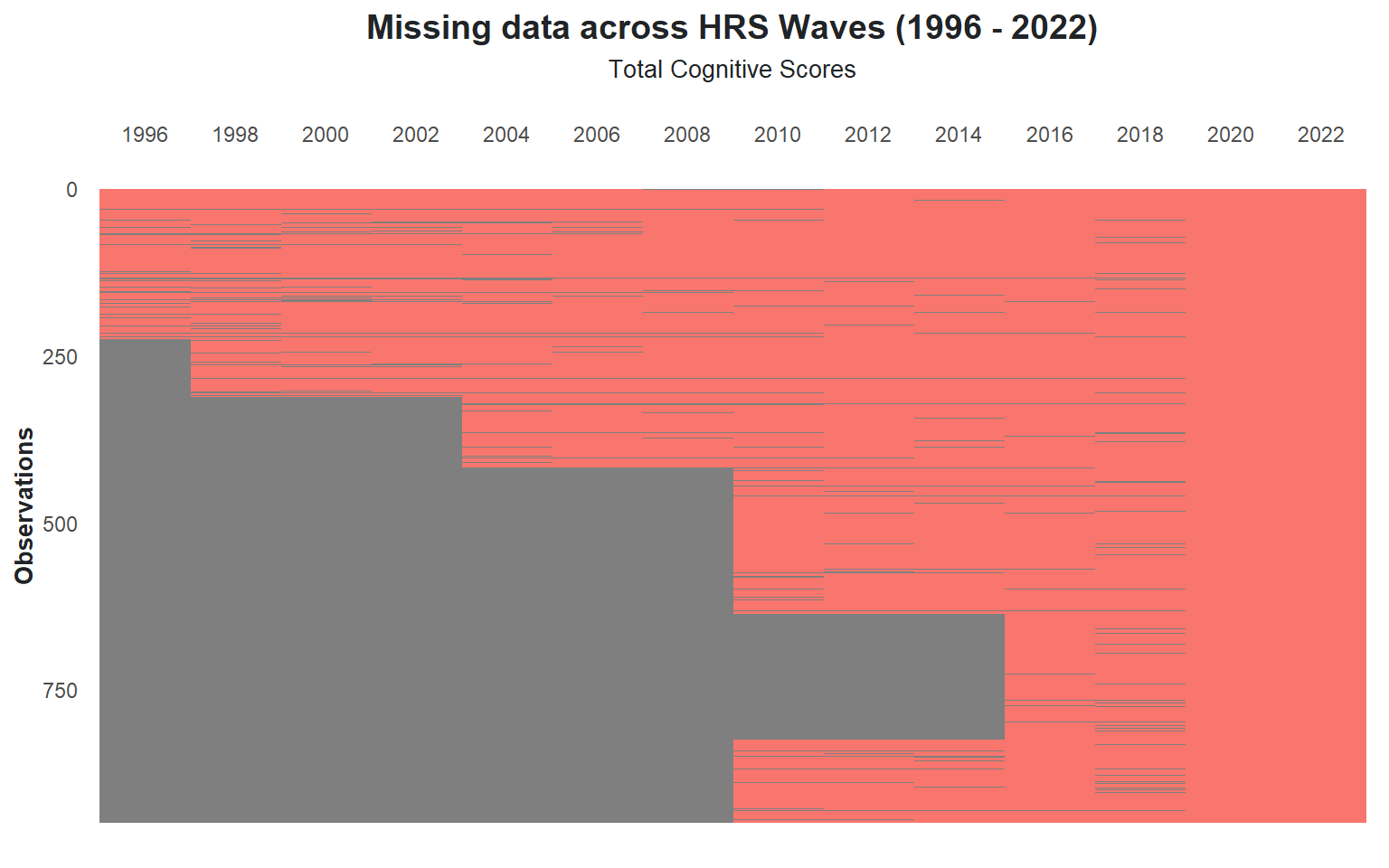
Since the participant pool is drawn from the 2020 wave of the HRS data, any analysis that looks back at previous waves will inevitably encounter missing data. Not all participants were included in earlier waves, leading to gaps in the data. @fig-missing-data displays the frequency of missing cognitive test data across each HRS wave.
```{r}
#| label: fig-missing-data
#| code-fold: true
#| code-summary: "Check out my code"
#| fig-width: 8
#| fig-cap: Occurance of missing data across HRS waves (participants were gathered from the 2020 HRS wave [in red])
#| warning: false
# Dynamic colours
colours <- c(rep("#2e2e2e", times = 12), "red", "#2e2e2e")
data |>
select(any_of(paste0("cogtot27_imp", 1996:2022))) |>
rename_with(~ str_replace(., "cogtot27_imp", ""), starts_with("cogtot27")) |>
visdat::vis_dat() +
labs(title = "Missing data across HRS Waves (1996 - 2022)",
subtitle = "Total Cognitive Scores") +
theme_clean() +
theme(legend.position = "none")
```
The HRS had its most recent participant intake in 2016, which explains the notable decline in missing data occurrences from that point onward. As new participants were not added after 2016, the dataset becomes more complete in subsequent waves, with fewer missing values.
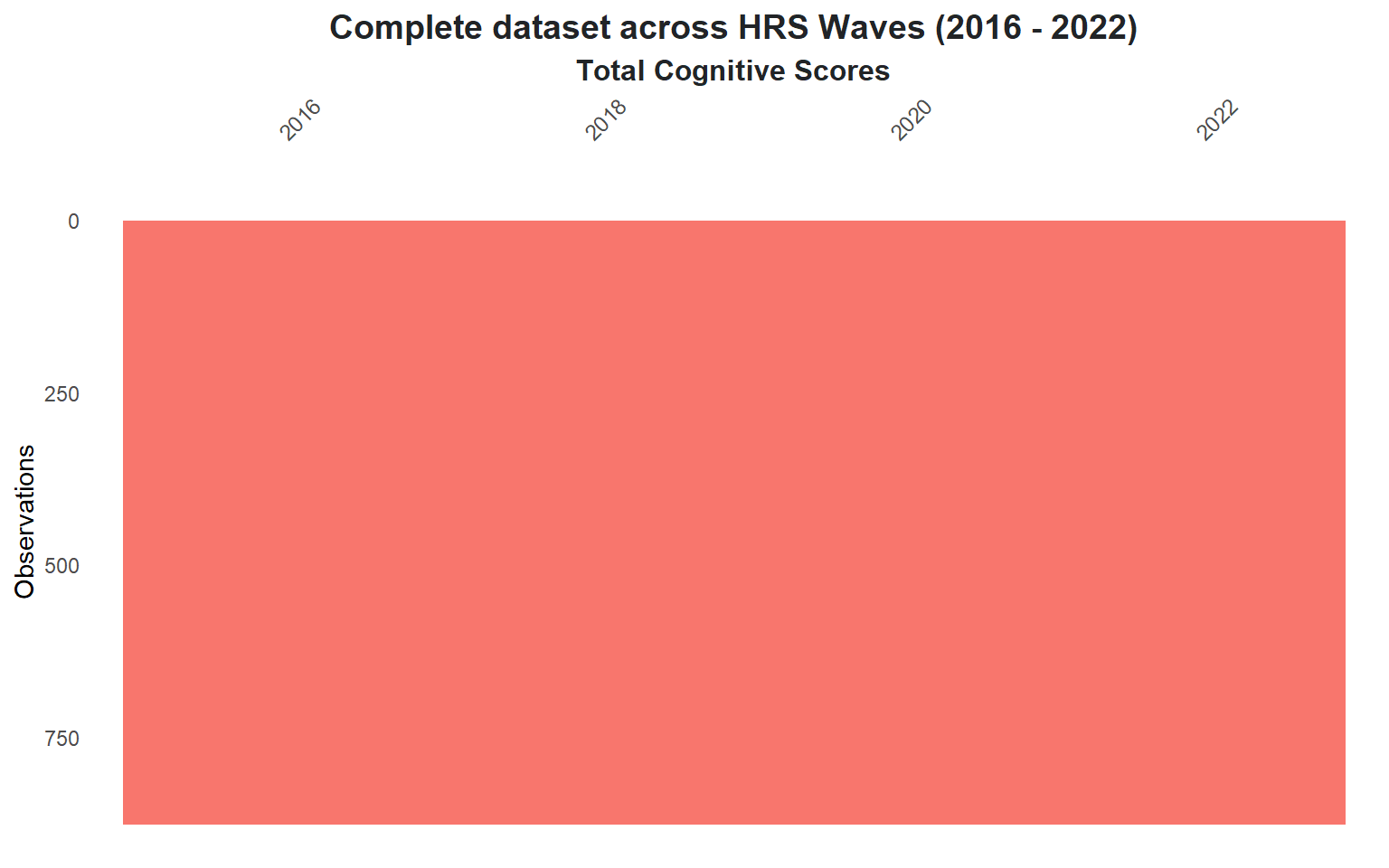
Given this shift, we will conduct our analysis **the reduced dataset (2016 - 2022)** (@fig-complete-data) along with removing the missing values from 2016 and 2018.
```{r}
#| fig-width: 8
#| code-fold: true
#| code-summary: "Check out my code"
#| label: fig-complete-data
#| fig-cap: HRS dataset after processing
#| warning: false
data |>
select(any_of(paste0("cogtot27_imp", 2016:2022))) |>
rename_with(~ str_replace(., "cogtot27_imp", ""), starts_with("cogtot27")) |>
na.omit() |>
visdat::vis_dat() +
labs(title = "Complete dataset across HRS Waves (2016 - 2022)",
subtitle = "Total Cognitive Scores") +
theme(
plot.title = element_text(
hjust = 0.5, size = 14, face = "bold", colour = colour),
plot.subtitle = element_text(
hjust = 0.5, size = 12, face = "bold", colour = colour),
axis.text.x = element_text(size = 9),
panel.grid = element_blank(),
legend.position = "none"
)
```
## Exploratory Data Analysis
### Cognitive Test Scores
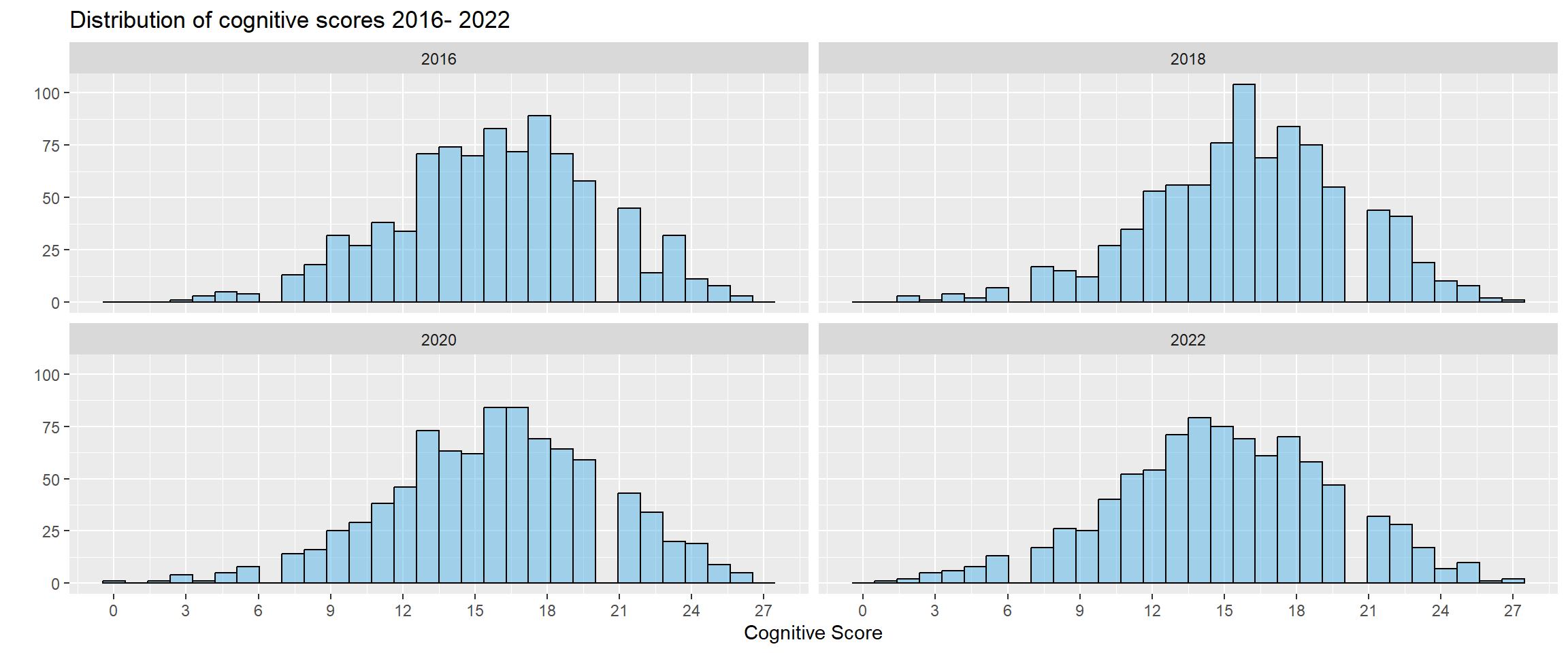
Initially, we will plot a distribution of the cognitive test scores (ranging from 0 - 27) across time for all HRS participants.
```{r}
#| message: false
#| fig-width: 12
#| label: fig-cognitive-test-scores
#| fig-cap: Cognitive test scores (2016 - 2022)
plot_cognitive_scores(data = data, year = 2016)
```
### Classification proportion per year
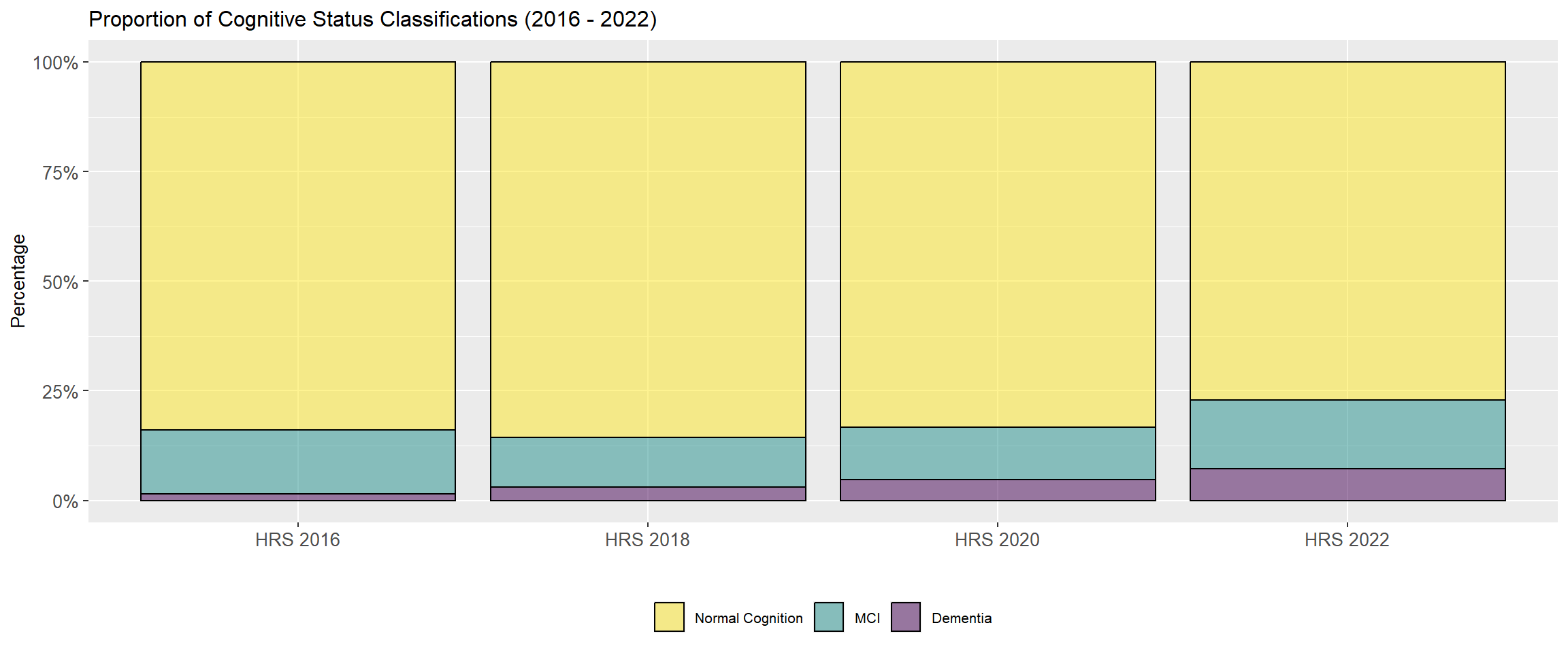
@fig-dementia-proportions shows the proportion of dementia classifications from HRS 2016 - 2022.
```{r}
#| fig-width: 12
#| label: fig-dementia-proportions
#| fig-cap: Proportion of dementia classifications per year
# Plotting proportions --------------------------------------------------------
data |>
count_transitions(years = seq(2016, 2022, by = 2), absorbing = TRUE) |>
ggplot(aes(x = Wave, fill = Classification)) +
geom_bar(position = "fill", alpha = 0.5, colour = "black") +
scale_y_continuous(labels = scales::percent) +
labs(title = "Proportion of Cognitive Status Classifications (2016 - 2022)",
x = "", y = "Percentage") +
scale_fill_viridis_d(direction = -1) +
theme(
axis.text = element_text(size = 10),
text = element_text(size = 10)) +
ggeasy::easy_move_legend("bottom") +
ggeasy::easy_remove_legend_title()
```
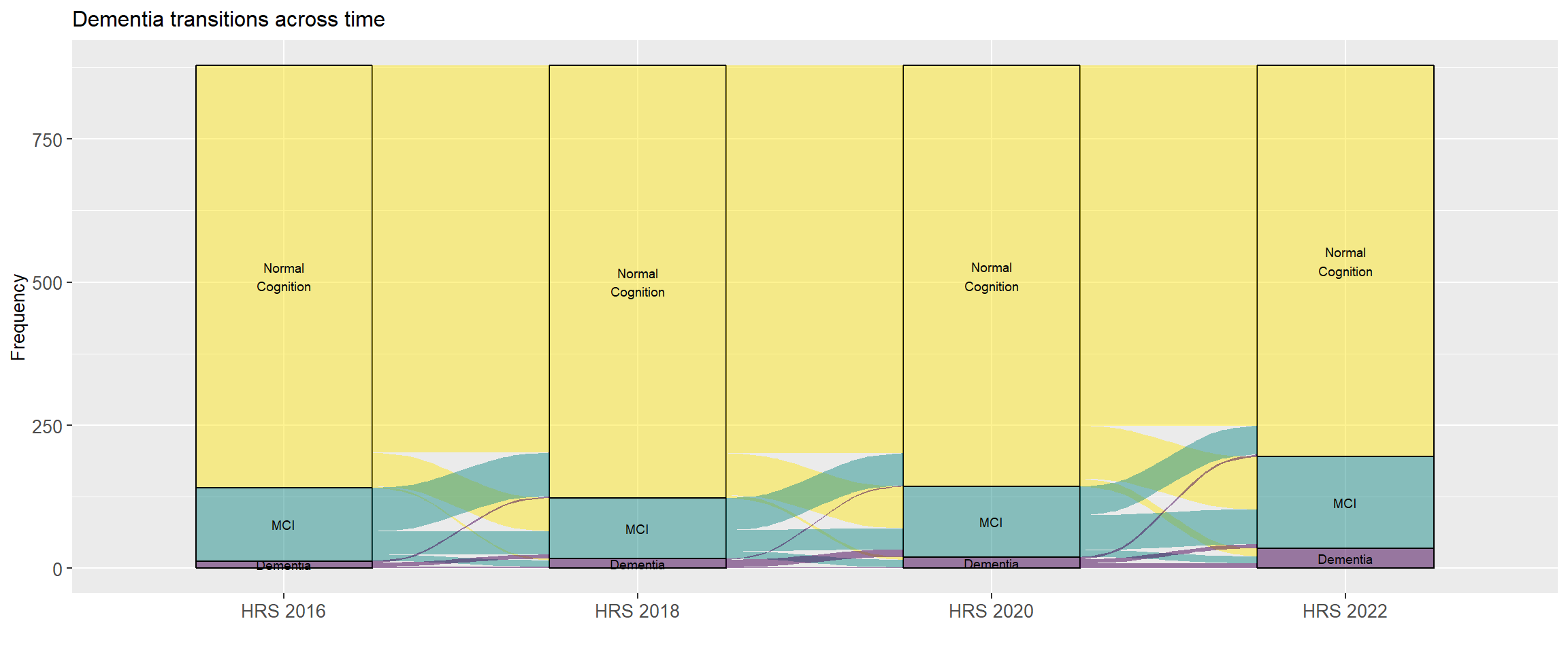
### Dementia transitions per year
@fig-dementia-transitions is an alluvial graph illustrating the transitions in cognitive classifications from one HRS wave to the next.
```{r}
#| fig-width: 12
#| label: fig-dementia-transitions
#| fig-cap: Dementia transitions (2016 - 2022)
data |>
count_transitions(years = seq(2016, 2022, by = 2)) |>
group_by(ID, Wave) |>
reframe(plyr::count(Classification)) |>
rename(Classification = x, n = freq) |>
plot_transitions(size = 2.5)
```
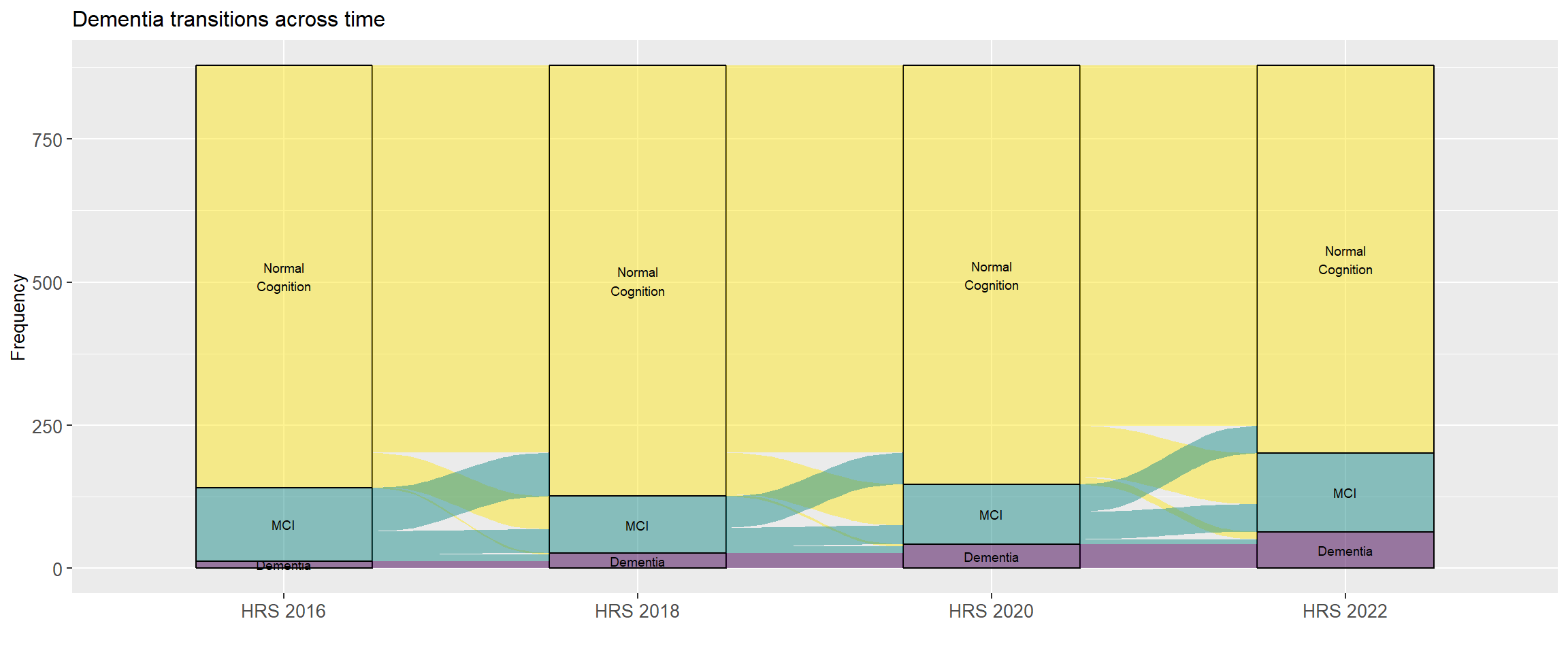
From this figure we can see that some individuals classified with dementia in one wave transition out of dementia in a subsequent wave. Let's fix this by adding an additional parameter to our `count_transitions()` function to turn dementia into an absorbing state (@fig-dementia-transitions-absorbing). We can do this by using the `cumany()` function from the `dplyr` [@Wickham2023] package.
```{r}
#| fig-width: 12
#| label: fig-dementia-transitions-absorbing
#| fig-cap: Dementia transitions with dementia as an absorbing state (2016 - 2022)
data |>
count_transitions(years = seq(2016, 2022, by = 2), absorbing = TRUE) |>
group_by(ID, Wave) |>
reframe(plyr::count(Classification)) |>
rename(Classification = x, n = freq) |>
plot_transitions(size = 2.5)
```